Converting raw files to mzML
FragPipe and MSFragger accept mzML files, which can be generated from your raw LC/MS data with MSConvert, part of the ProteoWizard software package. ProteoWizard can be downloaded here. Once ProteoWizard is installed, launch MSConvert.
This page has instructions for using MSConvert to generate mzML files from both Thermo Orbitrap and Bruker timsTOF data formats, though we recommend using the raw .d format for Bruker data.
Please note: The latest version of MSConvert should be used for TMT MS3 workflows. Philosopher is not compatible with mzML files generated using some older MSConvert versions, and the Windows taskbar may open an earlier version of MSConvert even if a more recent one is installed. Start MSConvert by clicking on the Windows icon, scrolling down to the desired (latest) version of ProteoWizard, and launching MSConvert program.
Please note: If you are using Linux, check the scripts in https://github.com/Nesvilab/msconvert-scripts
Convert Thermo .raw files (DDA or DIA data with non-staggered windows):
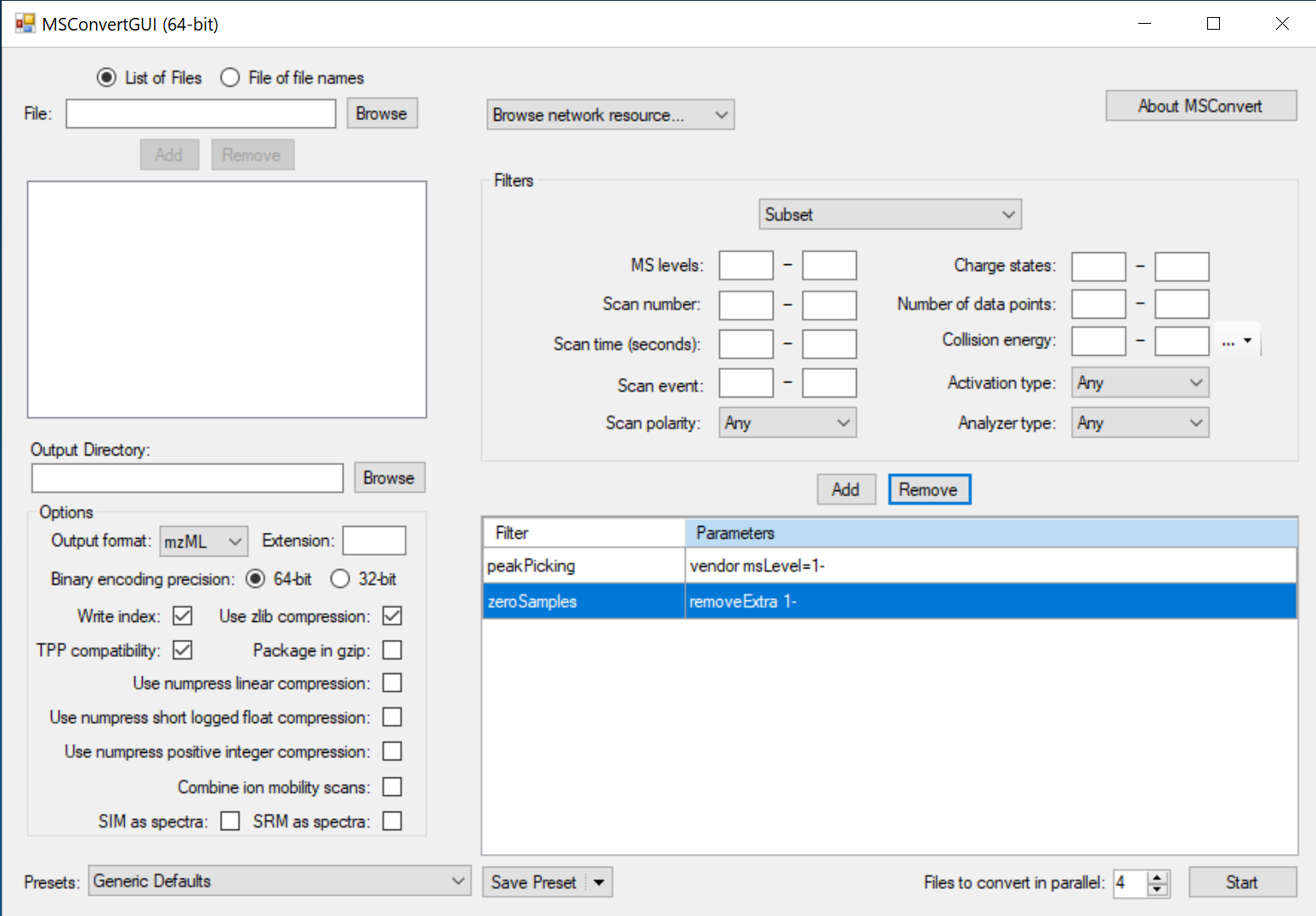
In the MSConvert GUI window:
- Browse for the raw files you want to convert.
- Add them to the input list.
- Select an output directory (where you want to mzML files to be generated).
- Set the output file format to ‘mzML’, and check the boxes for ‘Write index’, ‘TPP compatibility’, and ‘Use zlib compression’. Note: Do not select ‘zlib compression’ for use with Proteome Discoverer, which currently does not support this compression function.
- Select any additional filters you want to apply to the conversion. We recommend using the default filters shown here. “peakPicking” (centroiding) must be the first filter.
- Press ‘Start’ to begin converting your files.
Example command line options:
msconvert.exe --mzML --64 --zlib --filter "peakPicking true 1-" *.raw
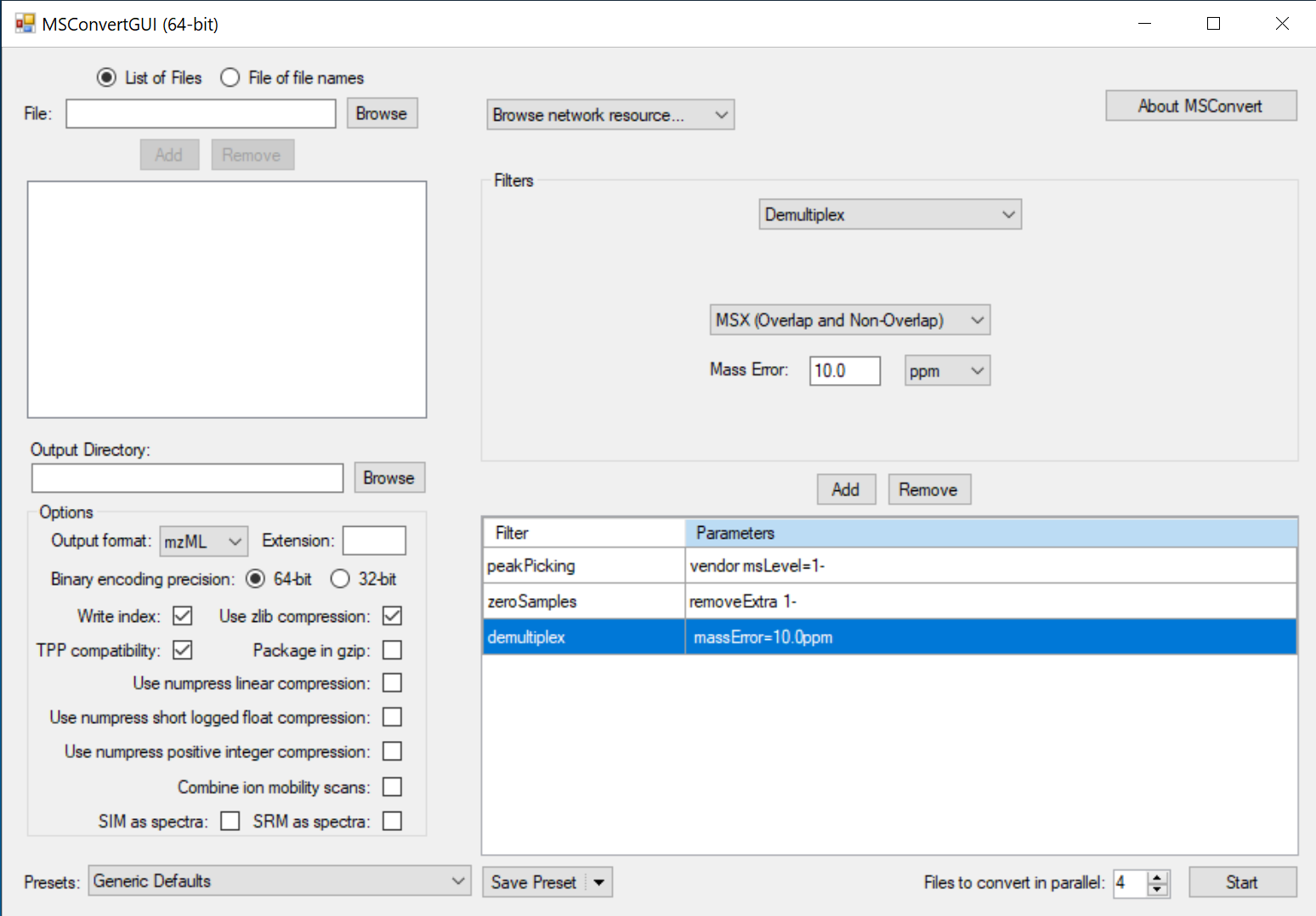
Convert Thermo DIA .raw files with staggered windows:
Note: “staggered” means the isolation windows have 50% overlapping. A small percentage of overlapping at the window boundary does not count.
Follow the steps above, with the addition of the “Demultiplex” filter in step 5.
Convert Bruker timsTOF .d files (NOT RECOMMENDED. MSFragger and IonQuant can load .d directly.):
In the MSConvert window:
- Browse for the .d folders you want to convert.
- Add them to the input list.
- Select an output directory (where you want to mzML files to be generated).
- Set the output file format to ‘mzML’, and check the box for ‘Combine ion mobility scans’. The boxes for ‘Write index’, ‘TPP compatibility’, and ‘Use zlib compression’ should also be checked.
- Add ‘peakPicking’ and ‘scanSumming’ filters.
- Press ‘Start’ to begin converting your files.
Note: with ‘scanSumming’ enabled, there is no ion mobility array in the converted file. Thus, the file is not suitable for MS1-based quantification.
