Running MSstats using MSstats.csv from IonQuant
IonQuant can generate a MSstats compatible file MSstats.csv.
Given an experimental setup that looks like this in the ‘Select LC/MS Files’ tab of FragPipe:
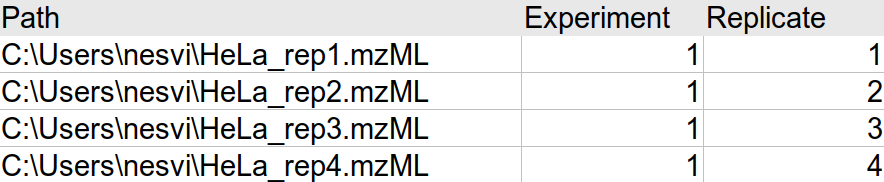
The MSstats.csv file output from IonQuant (via FragPipe) will look something like this:
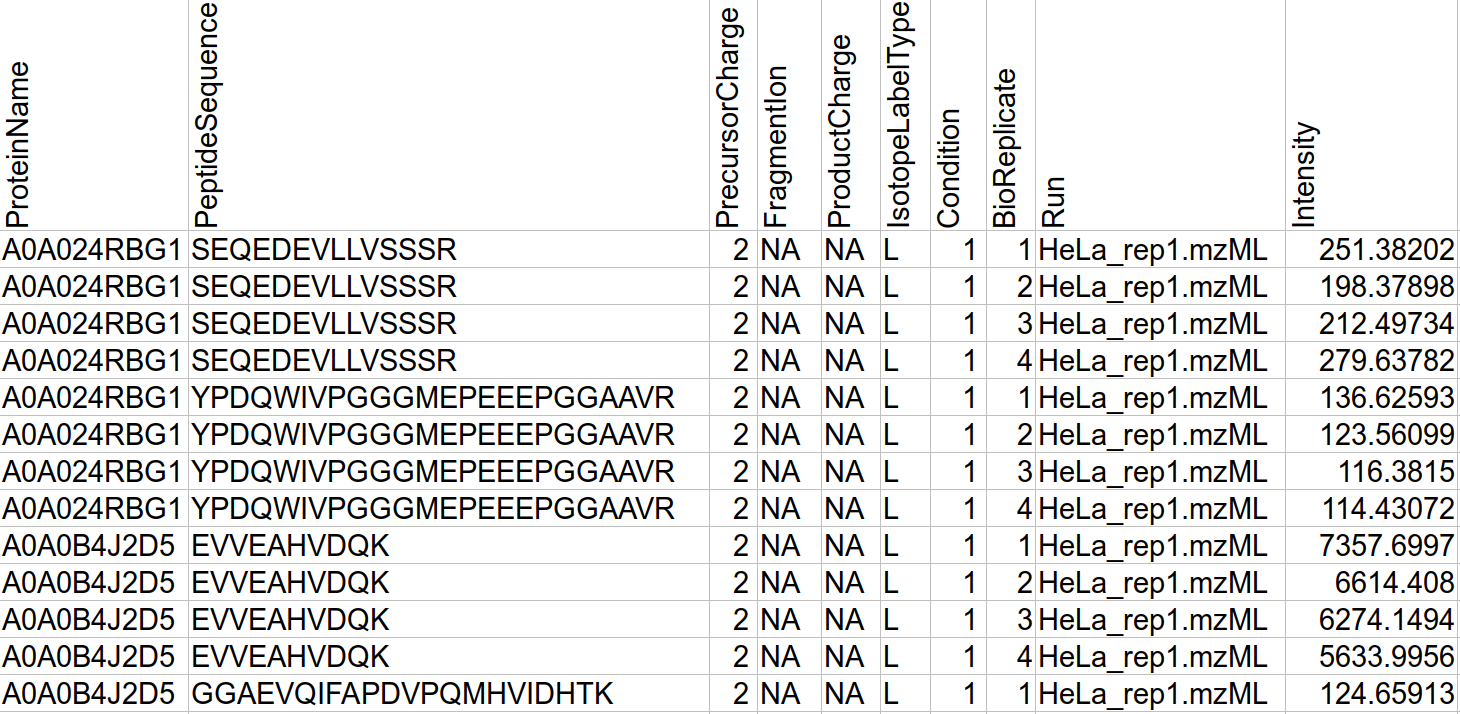
This MSstats.csv file can be read by MSstats without any conversion. The R command for installing MSstats is:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("MSstats")
Below is an example R script that reads and analyzes MSstats.csv using MSstats (make sure to set rootDir to the directory containing your MSstats.csv file):
rm(list = ls())
library(stringr)
library(readr)
library(MSstats)
rootDir <- "folder_with_MSstats.csv" # Specify the path of the directory containing MSstats.csv.
print(str_c("Using IonQuant's result from ", rootDir))
# Read MSstats.csv file.
raw <- read_csv(str_c(rootDir, "MSstats.csv"), na = c("", "NA", "0"))
raw$ProteinName <- factor(raw$ProteinName)
raw$PeptideSequence <- factor(raw$PeptideSequence)
# Change root directory for MSstats
print(str_c("Root DIR: ", rootDir))
setwd(rootDir)
# Processing the data using MSstats
processedData <- dataProcess(raw, logTrans = 10)
# Downstream analysis...
Details of the functions and options can be found here.
With processedData, users can perform various downstream analyses described in the MSstats user manual.