Setting up FragPipe
Install or update OpenJDK
FragPipe and MSFragger both require a 64-bit OpenJDK to run. Windows users can choose to download the -jre- version of FragPipe (see below) or install 64-bit pre-built OpenJDK here. Launch the installer and follow the prompts. You may need to restart FragPipe after updating OpenJDK.
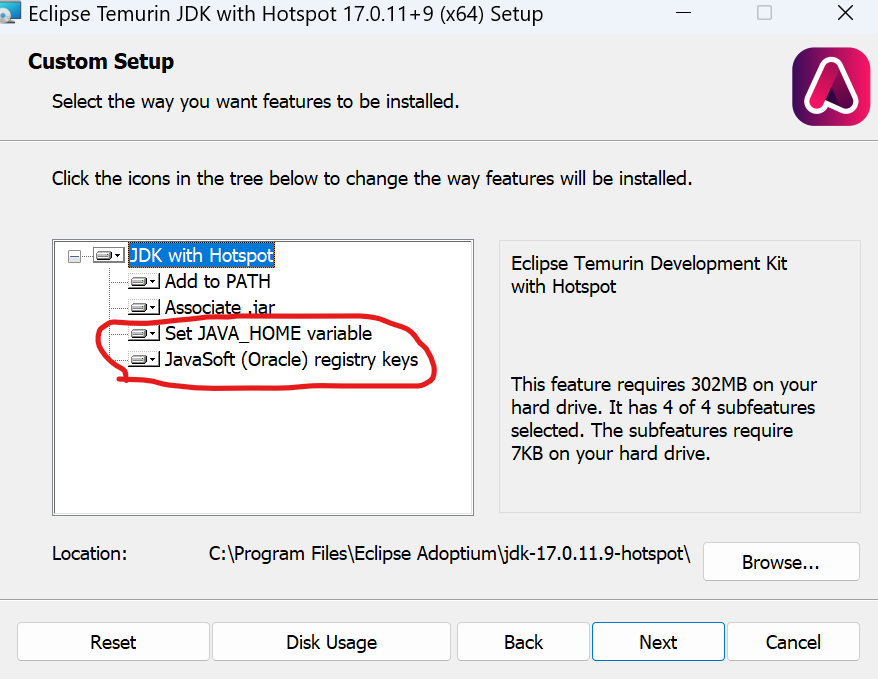
Note: During installation, remember to enable Set JAVA_HOME variable and JavsSOFT (Oracle) register keys.
Install Visual C++ Redistributable
Bruker .d reader and DIA-NN need Visual C++ Redistributable in Windows. If you see an error related to timsTOF data or DIA-NN, please try to install the Visual C++ redistibutable.
Install Mono (required for Thermo .raw file reading on Linux)
Linux users need to have Mono installed to read Thermo .raw files.
Install or update FragPipe
FragPipe can be downloaded here. Follow the instructions on that same Releases page to launch the program. When FragPipe launches, the first tab in the window (‘Config’) will be used to configure the program.
Optional: install, update, or use an already installed version of Python
Database splitting (to reduce the size of the in-memory fragment ion index– helpful for workstations with limited memory or for complex searches) and/or spectral library generation will require Python 3.
FragPipe requires Python 3.8 - 3.11
If you already have Python 3.8 - 3.11, specify the python executable file path in the Config tab, click Install/Upgrade EasyPQP, and wait for a few minutes.
If Python 3 is not already installed:
1) Download and install Python 3.11
Windows: download the installer from here
Linux: different Linux distributions have different commands to install Python. Please figure it out by yourself. If you are not familiar with the command line interface, please switching to Windows.
2) In FragPipe -> Config -> Python, use the ‘Browse’ button to navigate to the installation location and select python.exe. When FragPipe refreshes, DB Splitting and Spectral Library Generation should now be enabled.
3) To enable spectral library building with EasyPQP (which also works for timsTOF data), click Install/Upgrade EasyPQP at the bottom of the Config tab and wait for a few minutes. FragPipe will froze during the installation.