Labile Search in MSFragger and FragPipe
Labile PTM search extends mass offset/open searches for labile PTMs by capturing modification fragmentation in the MSFragger search. For details of the method and background, check out the MSFragger Labile paper at https://www.mcponline.org/article/S1535-9476(23)00048-8/fulltext.
This tutorial will provide an overview of the labile search methods and settings. To build a labile search workflow, we recommend loading of the of the built-in labile workflows in FragPipe (labile_phospho or labile_ADP-ribosylation) and adjusting the settings as needed.
If you are new to MSFragger or FragPipe searches, please first consult the Setup and Basic tutorials.
Tutorial contents
- Labile Search Overview
- MSFragger settings for Labile Search
- Validation Options for Labile Search
- Localization of Labile PTMs
- Quantification of Labile PTMs and Results
Labile Search Overview
Labile search is primarily a set of MSFragger options that allow modification fragmentation to be used in filtering and scoring spectra of modified peptides. There are 3 types of ions that we will refer to and 3 search types, outlined in the figure below.
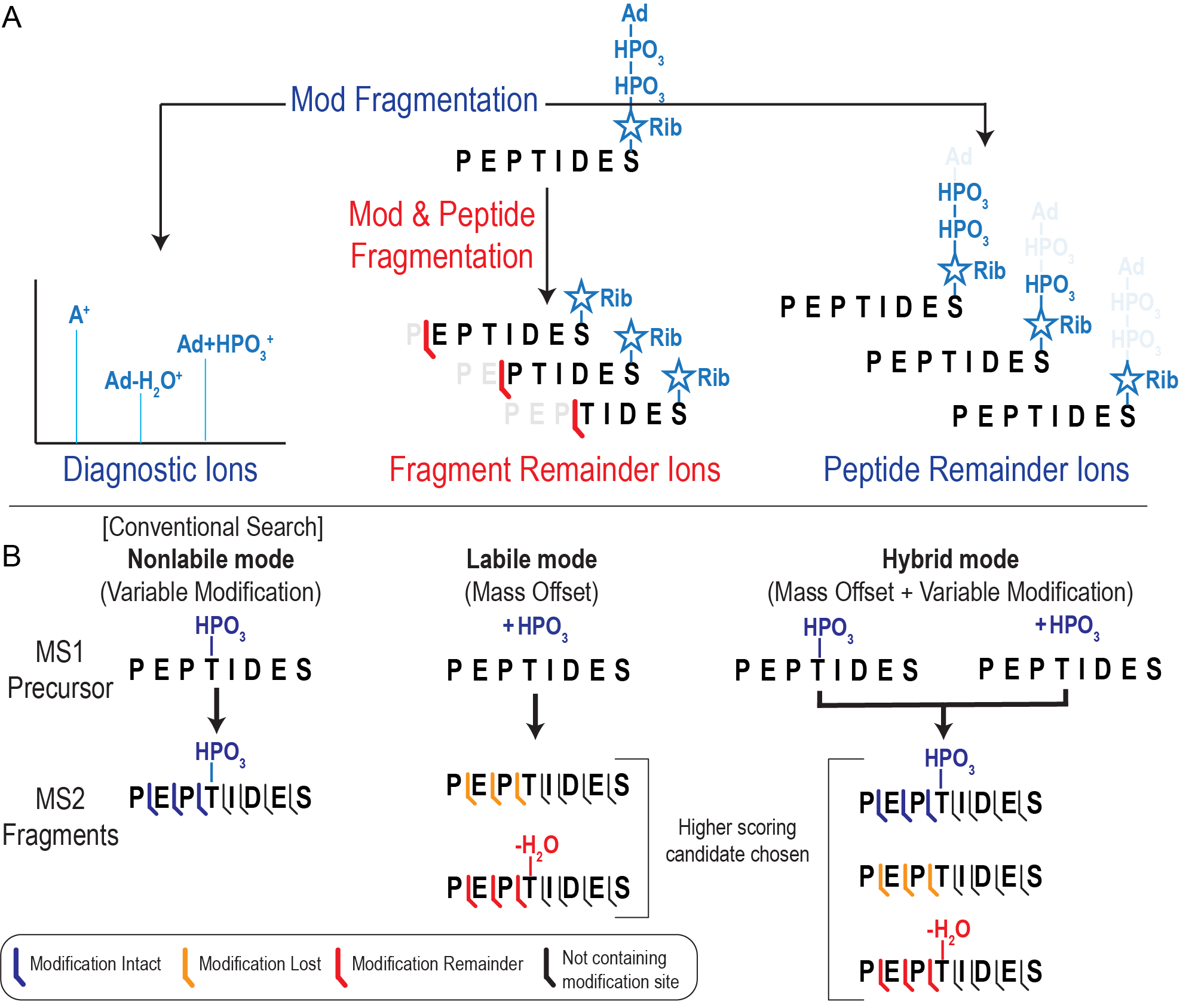
Diagnostic ions are only used for filtering which spectra are searched for delta masses (they
do not contribute to the score of a candidate PSM).
Peptide remainder ions are added to the hyperscore
of a candidate peptide when found.
Fragment remainder ions are added to the hyperscore AND used to
localize the modification (if localize_delta_mass is enabled).
If you are unsure of the fragment ions generated by a modification of interest, we recommend using the diagnostic ion discovery module in PTM-Shepherd to determine which ions to use for search.
Conventional Search (also called Variable Modification Search or Closed Search) only uses variable modifications
to search for PTMs, and does not support labile modifications.
Labile Search refers to using a mass offset
search to identify labile PTMs. Note that mass offset searches support only 1 delta mass (i.e., modification)
per peptide. Multiple labile modifications can be identified by adding mass offsets corresponding to
the mass of 2 or more modifications. Note that localization of modifications cannot always be accomplished
in these cases.
Hybrid Search searches for a modification as BOTH a variable mod and a mass offset. This
allows two things: 1) Partially labile modifications can be more easily detected as both intact and dissociated
forms of the modification are considered in the search. 2) Localization of multiple labile or partially labile
modifications can be accomplished when using only 1 mass offset (and encoding the remaining possible modifications
as a variable modifications).
MSFragger settings for Labile Search
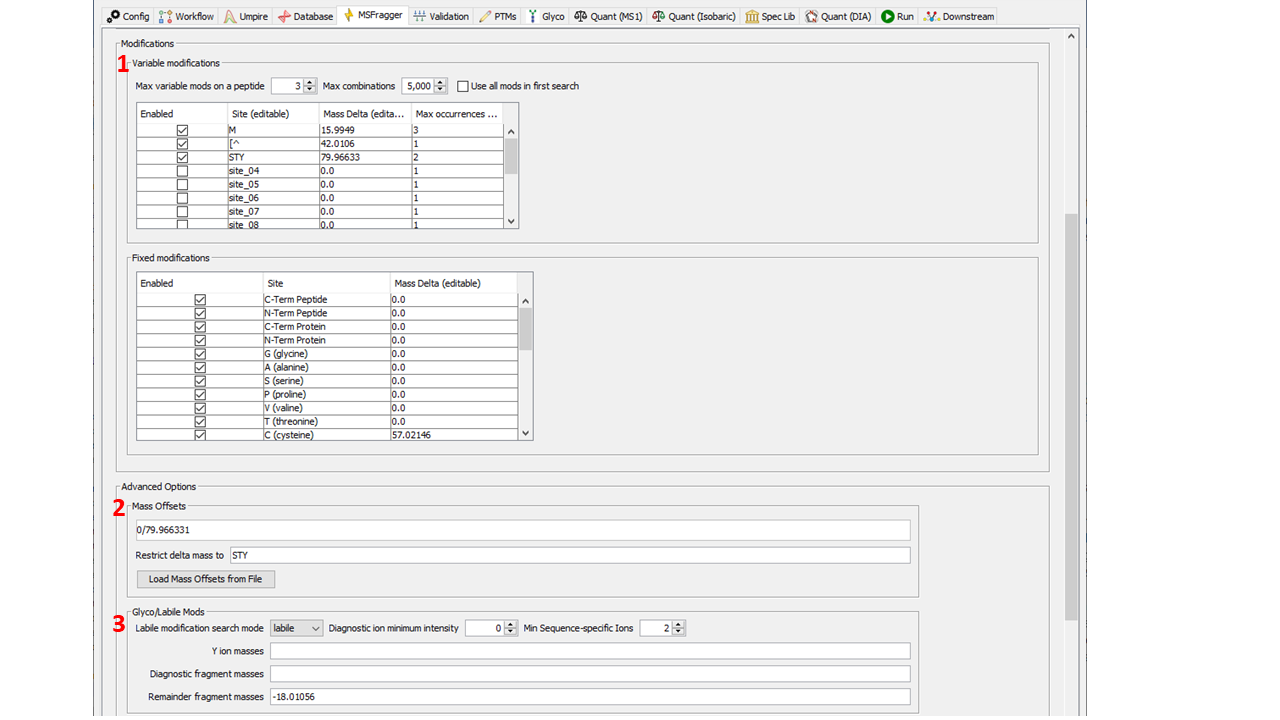
- Variable Modification settings. Include all nonlabile modifications here. Only include labile modifications if using a Hybrid search. The allowed amino acid residues, mass, and max number per peptide should be specified in the table.
- Mass Offset settings. Include all labile modifications here by listing their masses in the Mass
Offsets box. Mass offsets can be restricted to the amino acids listed in the box below, or allowed
on any amino acid using “all”.
NOTE: the restrictions are currently applied to ALL mass offsets equally. It is not yet possible to set specific amino acid restrictions for specific offsets.
NOTE: only 1 mass offset is considered per peptide, but combinations of labile modifications can be specified by adding their masses together.
NOTE: make sure to include 0 in the mass offset list! It is required for downstream processing (and even most enriched PTM datasets contain at least some unmodified peptides). - Labile Search Settings.
a) Use “labile” mode for labile searches.
b) Diagnostic ion minimum intensity: Threshold intensity ratio (between 0 and 1) to use for filtering spectra based on diagnostic ions. If a spectrum does not contain diagnostic ions, or their combined relative intensity is below this value, they will not be searched for mass offsets. c) Min sequence-specific ions: Peptide remainder ions are not sequence specific (as the peptide is not fragmented). This value is the minimum number of sequence specific ions required to report a candidate peptide (i.e., the minimum number of ions excluding peptide remainder ions). Mostly used for glycopeptide analyses. d) Y ion masses: Enter peptide remainder masses here (as neutral masses) e) Diagnostic fragment masses: Enter m/z values here (NOT neutral masses). f) Remainder fragment masses: Enter the masses of fragment remainders here (as neutral masses).
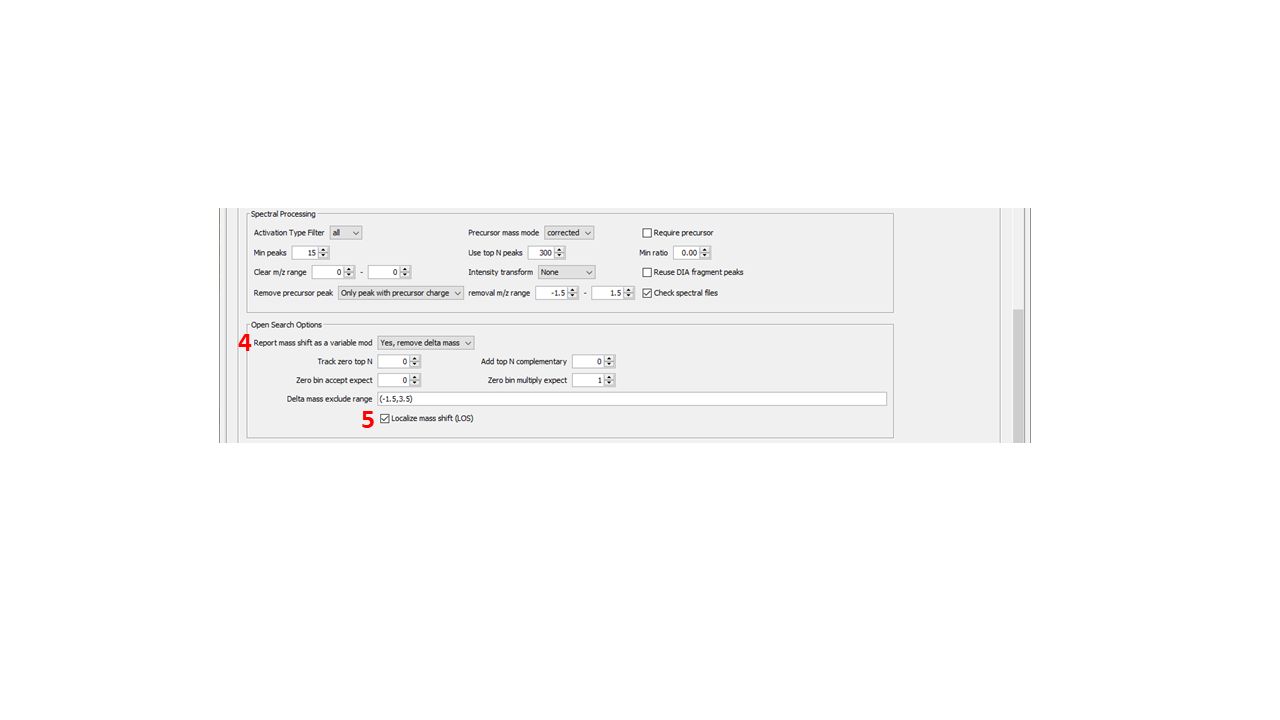
NOTE: adding fragment remainder masses greatly increases the number of fragment ions searched. It is recommended on to use highly prevalent remainder ions. - Mass Diff to Variable Mod. It is recommended to use this option (“Yes, remove delta mass”) for labile searches to have MSFragger report mass offsets in the same format as variable modifications in the output file for downstream validation and FDR. However, some searches (particularly where many different labile modifications are being considered) should not use this option. See additional discussion below in the Validation section.
- Localize mass shift (LOS). This option is REQUIRED to use fragment remainder masses. If no fragment remainder masses are specified, it can safely be turned off (it has no effect on a labile search unless fragment remainder masses are specified).
Validation Options for Labile Search
Either PeptideProphet or Percolator can be used for labile searches. Generally, Percolator offers slightly better performance, but can only analyze one MS data file at a time (whereas PeptideProphet can analyze all files together. This can be useful for improved statistics when looking for very rare PTMs). There are only a few cases where one tool is required over another: 1) Percolator. Is required when using MSBooster. Note that MSBooster supports a limited range of PTMs, so this is not usually an issue for PTM searches. 2) PeptideProphet. Is required if not using the “Mass Diff to Variable Mod” setting in MSFragger. If this is the case, use the extended mass model of PeptideProphet (load the “Mass Offset” settings in the PeptideProphet tab).
In all other cases, either tool can be used.
Localization of Labile PTMs
There are 2 options for labile PTM localization: MSFragger and PTMProphet.
- MSFragger Localization. MSFragger uses remainder fragment masses to localize labile (mass offset)
mods. This option works well when a single modification is present on a peptide and it leaves a clear
remainder fragment ion. Mass offset search will only consider a single localization site for the mass
offset - it will NOT attempt to decompose the offset into multiple sub-modifications and localize those.
Hybrid searches will also use the variable modification (conventional) localization method for intact
modifications.
NOTE: mass offset and hybrid searches require fragment remainder ions for localization. If no fragment remainder ions are specified, MSFragger will not perform any localization. - PTMProphet Localization. PTMProphet is an optional post-search localization that supersedes the MSFragger localization if it is run. It can be used to fine-tune localization of conventional, hybrid, and mass offset searches (as long as Mass Diff to Variable Modification is enabled). PTMProphet has recently added support for including neutral losses in the localization but encodes them differently from MSFragger. See the PTMProphet website for additional details.
Quantification of Labile PTMs and Results
If “Mass Diff to Variable Mod” is enabled in MSFragger, labile and hybrid searches can use label free or labeled quantitation tools in the same manner as conventional searches (no special adjustments are needed). See the various quantitation tutorials from the main FragPipe page for details.
Where to find PTMs in the Results tables
PTMs from labile search will be reported to the psm.tsv table in the Delta Mass column or the Assigned Modifications column if “Mass Diff to Variable Mod” is enabled in MSFragger. In the latter case, modifications will also be propagated to ion, peptide, and protein.tsv tables (and quant output tables, if applicable). Additional details about these tables can be found in the outputs tutorial.
Labile PTM visualization
The integrated FragPipe-PDV viewer can be used to inspect labile PTM spectra. See the viewer tutorial for further details.